High resolution 1 HLA matching has been shown to improve outcomes after haematopoietic progenitor cell transplantation (HPCT), reducing patient morbidity and mortality. A benefit to allelic resolution 1 genotyping is also becoming apparent in solid organ transplantation and transfusion, facilitating the definition of antibodies to specific eplets and allowing for the identification of acceptable (and unacceptable) HLA mismatches. These benefits apply across all the classical HLA loci, allowing the identification of e.g. permissive HLA-DPB1 mismatches in HPCT, and HLA-DR51/52/53 and HLA-DQA,DPA-directed antibodies in solid organ transplantation.
However, HLA typing has proven challenging due to the limited availability of serological reagents for phenotyping and the highly polymorphic nature of the HLA genes: some 18,504 alleles have been characterised for the classical HLA genes (IMGT/HLA release 3.33). Sanger sequencing has been the gold standard for high resolution HLA typing since the 1990s but this method is both costly and time-consuming: several rounds of testing are required to resolve ambiguities resulting from heterozygosity (cis/trans ambiguity) or limited gene coverage (e.g. null alleles caused by polymorphism outside of the peptide-binding groove-encoding exons). As a result of technological developments during the Human Genome Project, next-generation sequencing has evolved as an alternative approach for HLA genotyping.
Next-Generation Sequencing
Next-generation sequencing (NGS) is the massively parallel sequencing of clonal DNA molecules. NGS can be broken into three stages: library preparation, template preparation, and sequencing.
For HLA genotyping library preparation begins with targeted PCR amplification: this may involve long-range PCR of the entire HLA gene. The amplicons are then fragmented and adapters/indexes are introduced to facilitate subsequent templating and sequencing, and to allow the pooling of multiple ‘barcoded’ samples. Size selection is performed to select for fragments that will template efficiently yet also span (or ‘phase’ across) the majority of heterozygous positions, generating sufficient sequencing reads and preventing cis/trans ambiguity.
Template preparation and sequencing differ according to the NGS chemistry or ‘platform’ involved. For Illumina systems, library fragments affix to a flow cell where they undergo bridge amplification to generate clonal clusters; these are then denatured prior to sequencing. Illumina utilise ‘sequencing by synthesis’ and fluorescent detection: differentially-labelled dNTPs simultaneously flow across the flow cell and the emission spectrum of each cluster is captured following incorporation. Adapter-ligated fragments are sequenced in both directions (‘paired-end sequencing’). Ion Torrent systems bind and clonally-amplify fragments on Ion sphere particles (ISP) which are then loaded onto an Ion chip, such that one ISP is captured per well. dNTPs sequentially flow across the chip and ‘semiconductor sequencing’ captures dNTP incorporation via the release of a proton, causing a pH change and voltage in that specific well. ISP-bound fragments are sequenced in a single direction (single read sequencing). Alternative platforms for template preparation and sequencing include nanopore sequencing (currently limited for HLA genotyping due to the high sequencing error rate) and ‘single molecule real-time sequencing technology’ (SMRT) by Pacific Biosciences (PacBio). SMRT utilises a circular template, allowing for the sequencing of longer reads: fluorescently-labelled dNTPs are incorporated and fluorescence is emitted as the label is cleaved.
Finally, alternate samples must be disentangled using the indexes (barcodes) inherent in each fragment, and the sequence of the targeted gene reconstructed using bespoke software.
Clinical Utility
Next-generation sequencing is able to provide first-pass high resolution HLA genotyping (and in many cases allelic resolution genotyping) with a very low ambiguity rate. Several publications describe the validation of NGS for clinical HLA genotyping: on the Illumina MiSeq 2 and the Ion Torrent PGM platform 3. NGS provides high accuracy and precision (reproducibility) and offers a significant cost-saving relative to Sanger (not least with respect to labour 3 4 ). Guerra et al. (2018) 3 also demonstrate that high-resolution genotyping by NGS can be achieved within acceptable turnaround times for a clinical laboratory.
Like many methodologies, the quality of the HLA NGS output is contingent on the quality of the reagents and analysis software. Allelic ambiguity can result from incomplete gene coverage in the initial PCR amplification, and genotypic ambiguity may result if fragment size is insufficient to span (phase) two heterozygous positions. HLA-DPA1, -DPB1 and -DQB1 have proven particularly difficult to sequence 2 3, and phase ambiguities typically occur for HLA-DPB1 due to the genetic distance between exons I and II. The input quantity of DNA is also critical (dsDNA must be quantified by e.g. Qubit or an alternative fluorometric system) and can be problematic in patients who are cytopenic, or from alternative sample sources (for example, buccal cell or saliva samples).
AllType™ NGS
One Lambda bring their HLA expertise to NGS in the form of AllType™: a NGS workflow that begins with the PCR amplification of all 11 classical HLA genes (HLA-A,B,C,DRB1,DRB3/4/5,DQA1,DQB1,DPA1,DPB1) in a single multiplexed reaction. This reduces the quantity of DNA required to only 50 ng (whilst alternative products require up to 1500 ng), and additionally decreases consumable (and third-party reagent) usage, margin for error, and handling time. 24 samples are handled across 24 wells as opposed to 216 wells (three 96-well plates) for some products, reducing the need for automation. An initial thermal cycling time of only 2.5 hours means that AllType™ is the only NGS workflow that can be completed on the bench in a single workday; samples can be ready for analysis within two workdays.
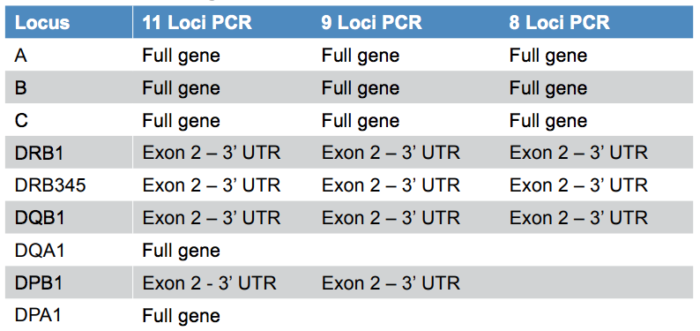
AllType™ NGS minimises allelic ambiguity by offering excellent gene coverage (Table 1): whole gene for HLA-A,B,C,DQA1,DPA1 and exon II through 3’ UTR for HLA-DRB1,DRB3/4/5,DQB1,DPB1. The AllType™ NGS Amplification Kit is also available in 11, 9 and 8 Loci formats, allowing user flexibility.
Table 1: Gene content and coverage for the AllType™ NGS Amplification Kit.
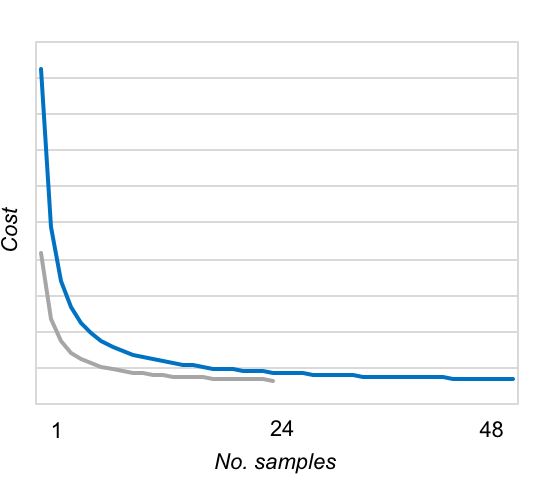
The AllType™ NGS workflow is facilitated by a user-friendly workbook-based calculation tool and requires minimal upfront investment (additional to a Qubit or other system for dsDNA quantitation, and access to an Illumina MiSeq or Ion Chef™ and Ion GeneStudio™ S5 system). In contrast to its predecessor (NXType™ NGS), AllType™ NGS is supported on both Illumina and Ion Torrent platforms, with the Ion Chef™ automating template preparation and Ion chip loading for the Ion Torrent workflow. Up to 48 samples can be processed in a single sequencing run, although fewer samples can be processed with the workflow remaining cost-effective once a threshold number of samples has been reached (Figure 1).
Figure 1: Next-generation sequencing rapidly becomes cost effective using either Illumina Standard (blue) or Micro (grey) flow cell, or the Ion Torrent Ion 530™ Chip (blue).
Finally, AllType™ NGS received CE-marking for in vitro diagnostic testing (CE-IVD) in May 2018 for the 11-Loci Amplification Kit and the accompanying TypeStream Visual™ NGS analysis software, demonstrating the quality and robustness of One Lambda’s NGS workflow.
TypeStream Visual™ Analysis Software
TypeStream Visual™ is intelligent software for the analysis of AllType™ NGS assays. In contrast to the browser-based TypeStream™ plugin (now discontinued), TypeStream Visual™ is a standalone software solution able to analyse both single read (Ion Torrent) and paired-end (Illumina) sequencing data. Sequencing ‘reads’ are mapped against an IMGT/HLA reference library to determine the HLA genotype. Additionally, a wide range of analytical tools, run statistics and quality metrics are available to reinforce decision making. Auto-analysis of Illumina FASTQ and Ion Torrent BAM files following sequencing further reduces the time to results, maximising the efficiency of the AllType™ NGS workflow. TypeStream Visual™ software features a familiar ‘front-end’ environment (Figure 2), allowing HLA typing integration with HLA Fusion™ Research and associated functionality.
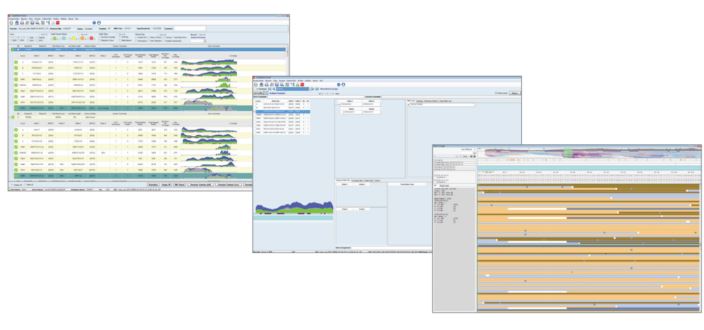
Figure 2: TypeStream Visual™ software presents stratified data, allowing the user both clarity of results and in-depth interrogation of sequencing data.
AllType™ NGS and TypeStream Visual™ build upon One Lambda’s existing range of HLA genotyping products, allowing user choice from presence-absence testing through to allelic resolution genotyping. AllType™ NGS is an exceptional next-generation sequencing product suitable for both low and high throughput laboratories. View the One Lambda webinars 5 6 and contact the VH Bio team for further information.
References
- Nunes, E., Heslop, H., Fernandez-Vina, M., Taves, C., Wagenknecht, D.R., Eisenbrev, A.B., Fischer, G., Poulton, K., Wacker, K., Hurley, C.K., Noreen, H. and Sacchi, N. (2011). Definitions of histocompatibility typing terms. Blood, 118(23): 180.
- Weimer, E.T., Montgomery, M., Petraroia, R., Crawford, J. and Schmitz, J.L. (2016). Performance characteristics and validation of next-generation sequencing for human leucocyte antigen typing. The Journal of Molecular Diagnostics, 18(5): 668.
- Guerra, S.G., Chong, W., Brown, C.J. and Navarrete, C.V. (2018). Evaluation of Ion Torrent sequencing technology for rapid clinical human leucocyte antigen typing. International Journal of Immunogenetics, 45(4): 230.
- Stoddard, J.L., Niemela, J.E., Fleisher, T.A., Rosenzweig, S.D. (2014). Targeted NGS: a cost-effective approach to molecular diagnosis of PIDs. Frontiers in Immunology, 5: 531.
- One Lambda Webinar: Validation of HLA Typing by NGS (Recorded 2018 May 23). http://www.onelambda.com/en/knowledge-support/Education/webinars.html
- One Lambda Webinar: Analyzing One Lambda’s AllType NGS Assay using TypeStream Visual Software – Version 1.2 (Recorded 2017 Jun 22). http://www.onelambda.com/en/knowledge-support/Education/webinars.htm
